There are an abundance of “scientific workflow” frameworks, job schedulers, queuing systems, MPI implementations, and language-level primitives for multiprocessing. Sometimes, though, you just have to do a task a bunch of times. Here, I introduce GNU parallel for a simple task and a more advanced task.
Convert files to png
Maybe you just rendered a bunch of images with VMD. You try to insert them into your powerpoint presentation, but the files are tga. You can convert them with ImageMagick’s convert program. Instead of running this utility by hand each time, you can do some fancy command-line trickery:
find . -name "*.tga" -exec convert {} {}.png \;
This works, but can be improved with parallel
find . -name "*.tga" | parallel convert {} {.}.png
This method offers two advantages:
This will run the conversion task in parallel, exploiting your multi-core CPU
The new filenames will end in
.pnginstead of.tga.png
Run a grid search over parameters
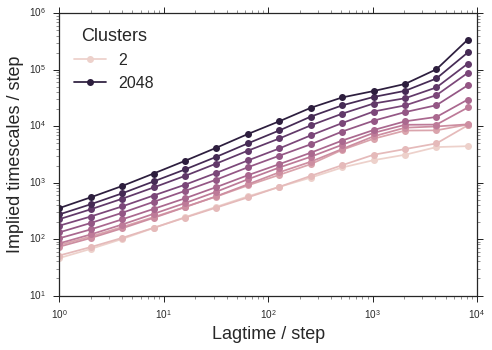
tICA is a powerful algorithm for discovering “reaction coordinates” from molecular dynamics datasets. It’s a linear algorithm, but recent advances show how the kernel trick can be used to learn non-linear coordinates. Approximations will probably work well here. Suppose we want to calculate the tICA timescales over a grid of (lag-time, degree-of-approximation) parameter values. This is “embarrassingly parallel”. We can write our script to exploit some more advanced features of parallel.
$ parallel echo ::: `seq 1 2` ::: a c
1 a
1 c
2 a
2 c
parallel will automatically do the “outer product” of argument lists, separated by :::. Below, we use one script to both generate and consume argument lists. This script uses MSMBuilder to fit multiple tICA models.
#!/usr/bin/env python
# Usage:
# parallel ./tica.py ::: `./tica.py --1` ::: `./tica.py --2`
n_clusters = [2 ** i for i in range(1, 13)]
lag_times = [2 ** i for i in range(14)]
import sys
if sys.argv[1] == '--1':
for nc in n_clusters:
print(nc)
sys.exit(1)
if sys.argv[1] == '--2':
for lt in lag_times:
print(lt)
sys.exit(2)
nc = int(sys.argv[1])
lt = int(sys.argv[2])
components = min(10, nc)
from msmbuilder.io import load_trajs, save_generic
from msmbuilder.decomposition import tICA
# Load input trajectories, precomputed
# These have different numbers of features
meta, ftrajs = load_trajs("ftrajs-{}".format(nc))
tica = tICA(n_components=components, lag_time=lt, kinetic_mapping=True)
tica.fit(ftrajs.values())
res = {'lagtime': lt, 'clusters': nc}
for i in range(components):
res['timescale_{}'.format(i)] = tica.timescales_[i]
save_generic(res, 'tica-{}-{}.pickl'.format(nc, lt))
I also include code for combining the results into one dataset in the same script by adding the additional clause to argument parsing:
import pandas as pd
from msmbuilder.io import load_generic
if sys.argv[1] == '--combine':
results = []
for nc in n_clusters:
for lt in lag_times:
results += [load_generic('tica-{}-{}.pickl'.format(nc, lt))]
df = pd.DataFrame(results)
df.to_pickle('tica-timescales.pandas.pickl')
sys.exit(3)